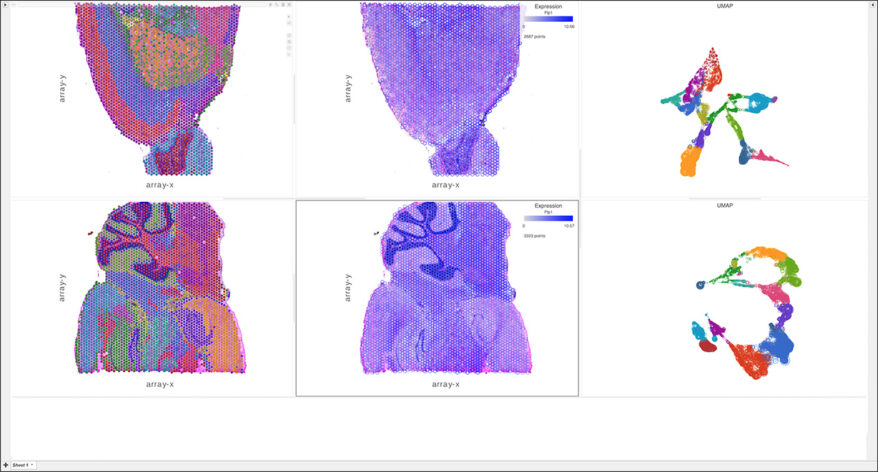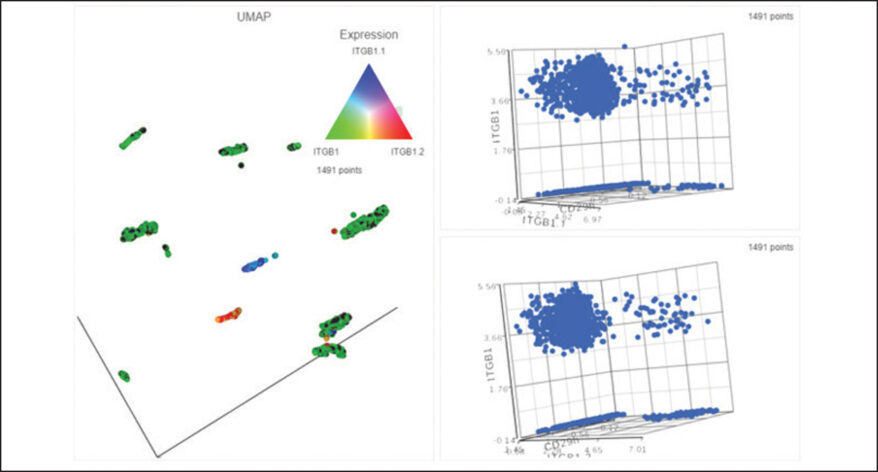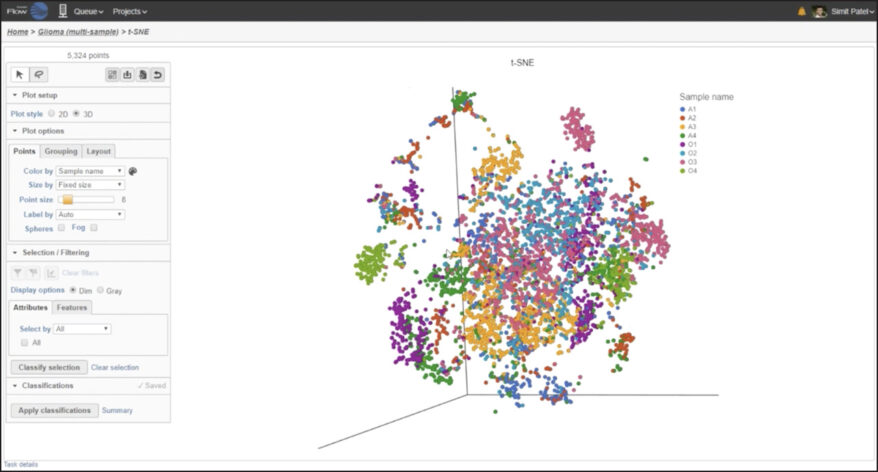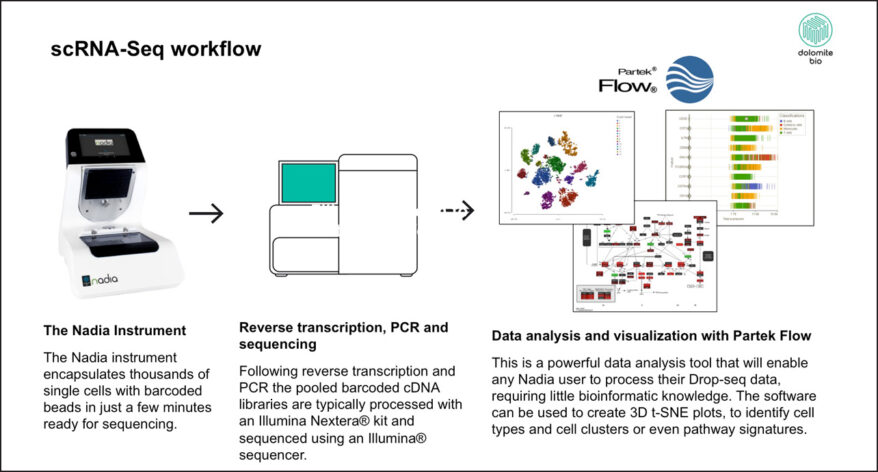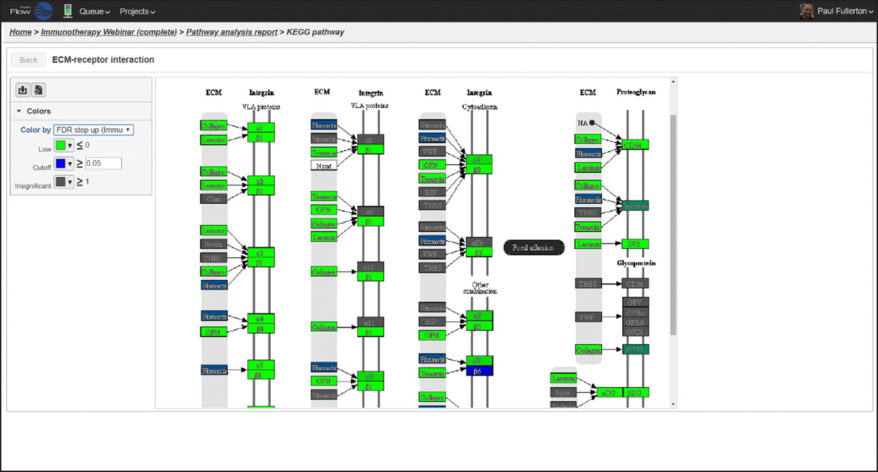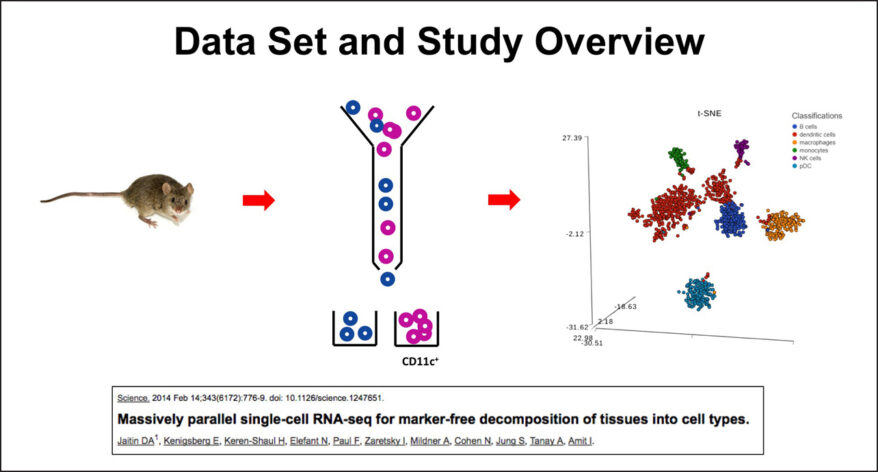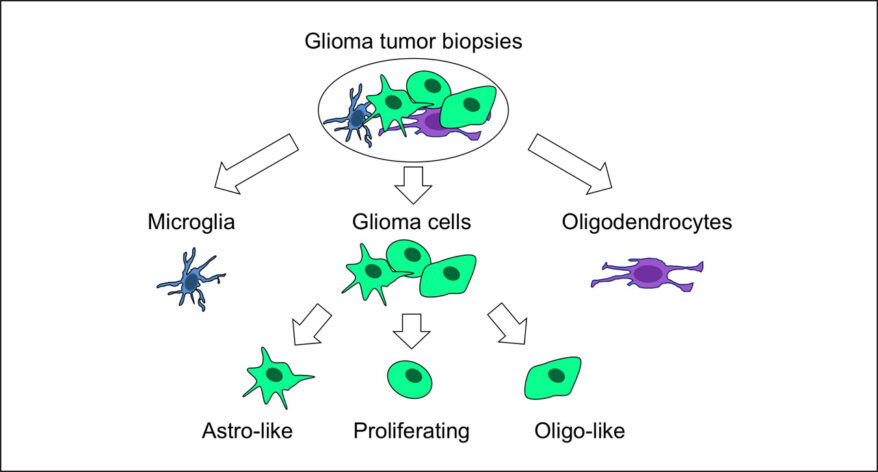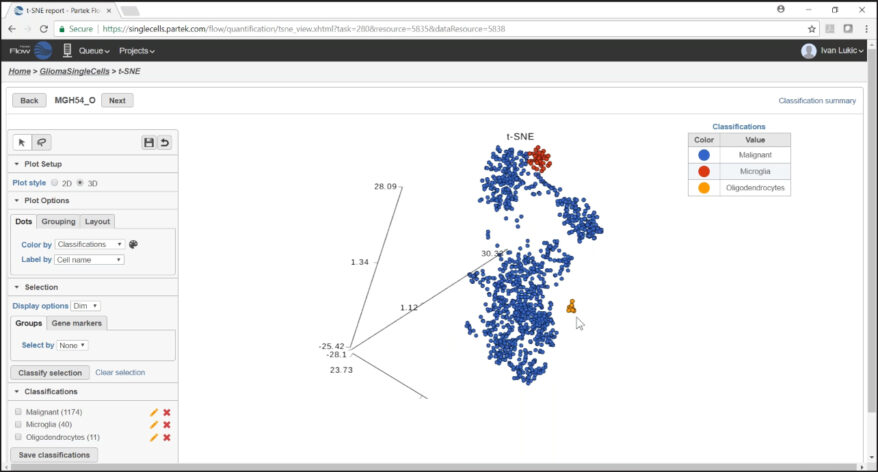
New Single Cell and Bulk RNA-Seq Features of Partek Flow Software
We have been adding new features to Partek Flow bioinformatics software to bring even more speed, functionality, and ease-of-use to your single cell and bulk gene expression studies. Join us to learn about these features and how they can be applied in your data analysis.
Features being discussed:
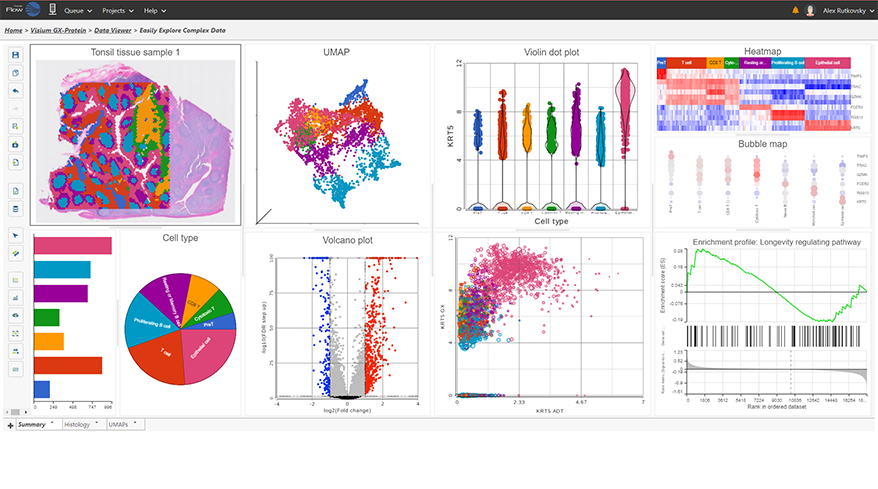
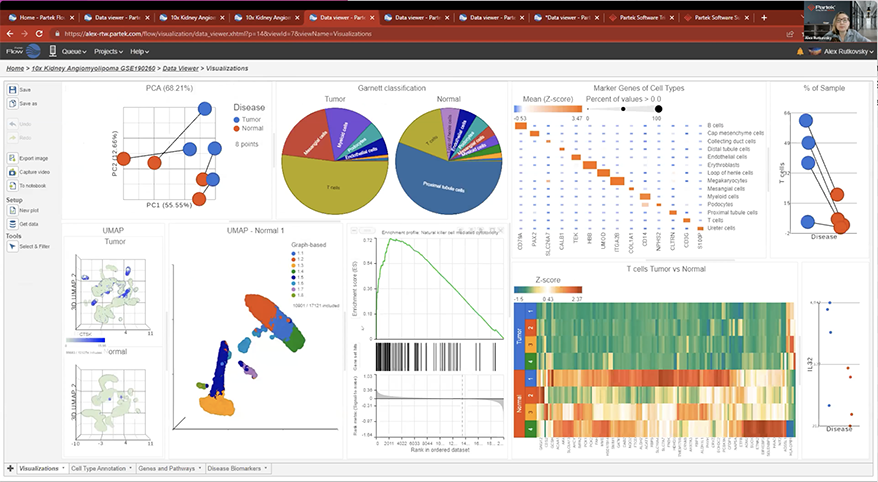
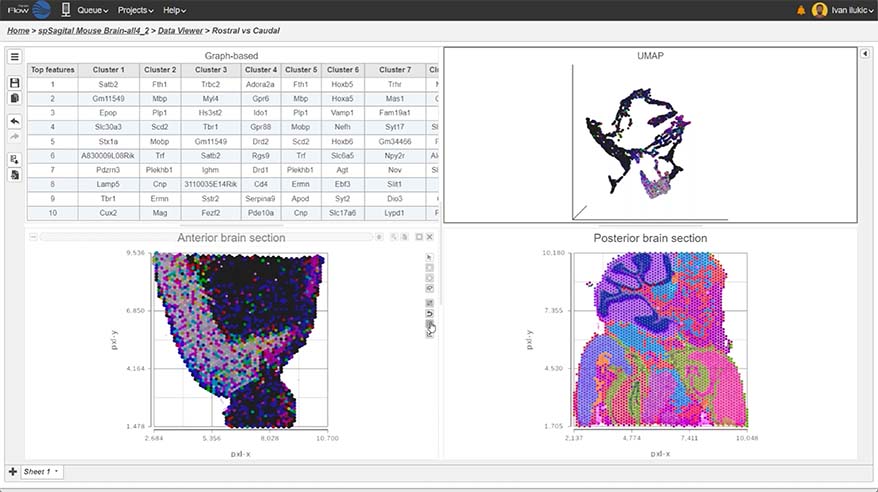
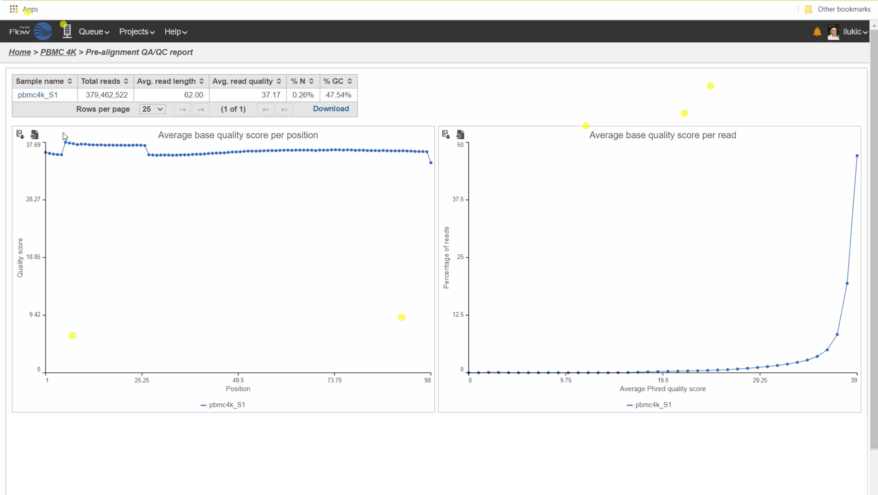
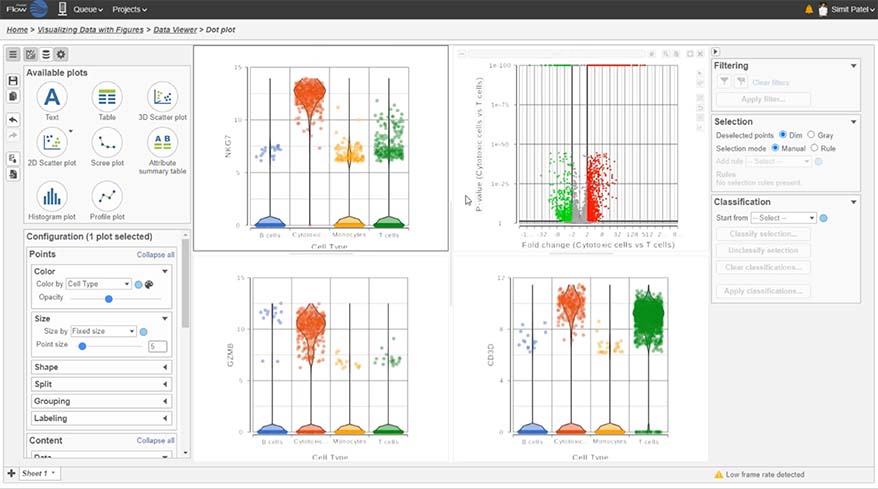
- New Ribosomal QAQC plot for single cell data
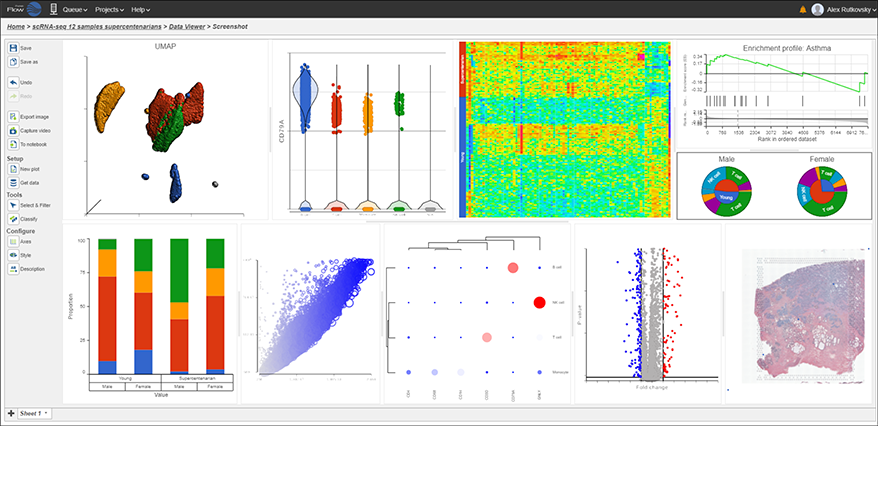
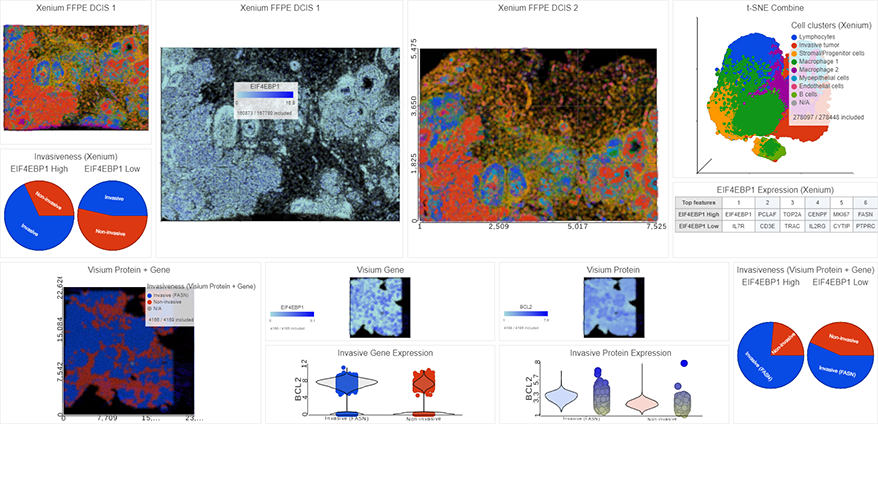
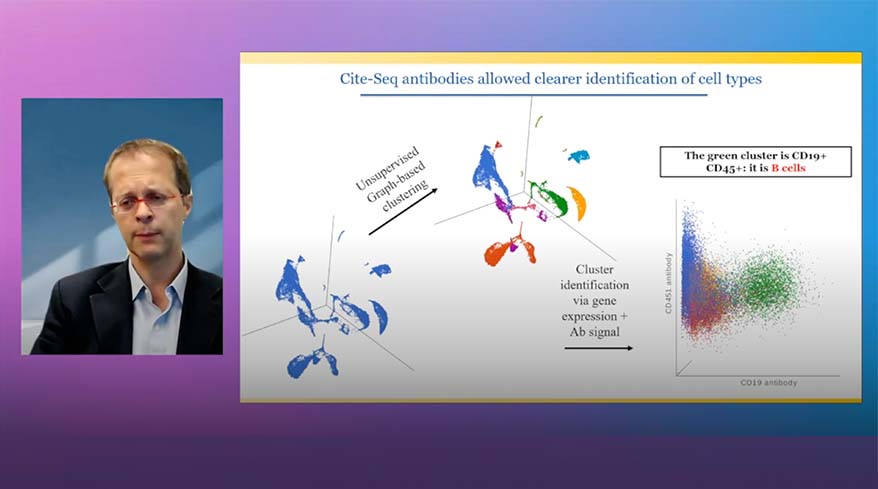
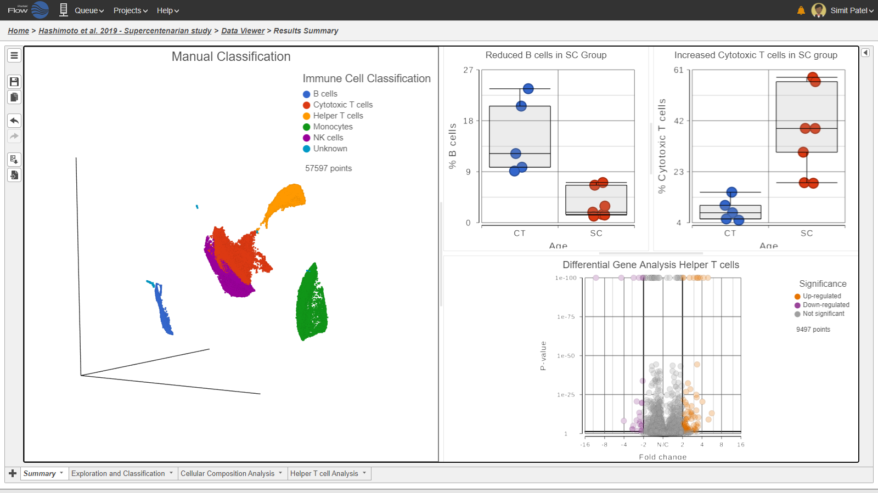
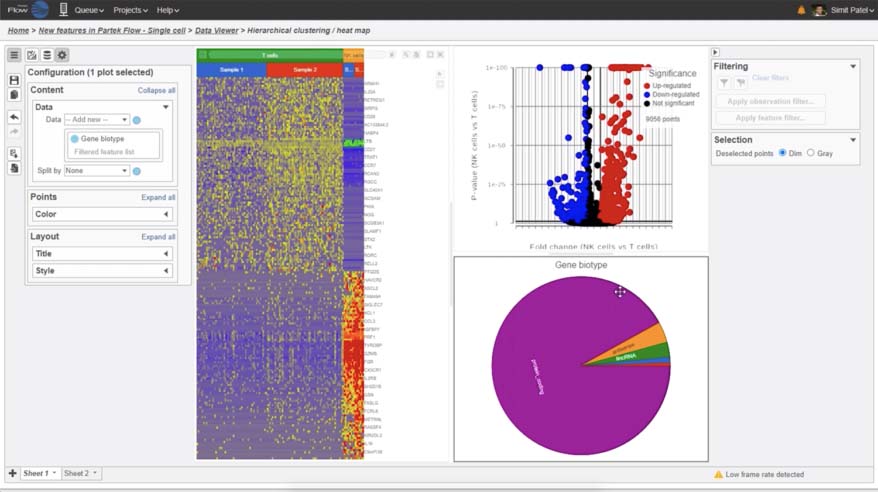
- Improved heatmap and volcano plots in visualizing single cell and bulk RNA-Seq data
- New pie charts
- Improved DESeq2 for differential gene expression
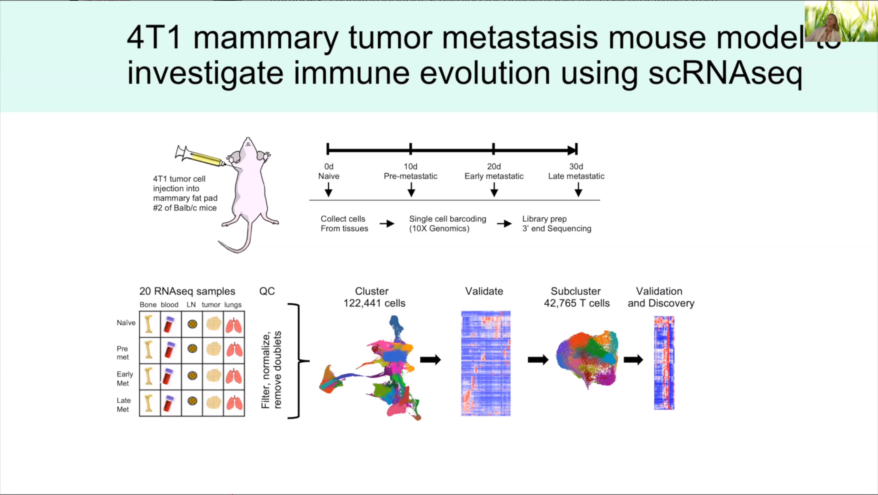
- New Scran and SCTransform normalization methods
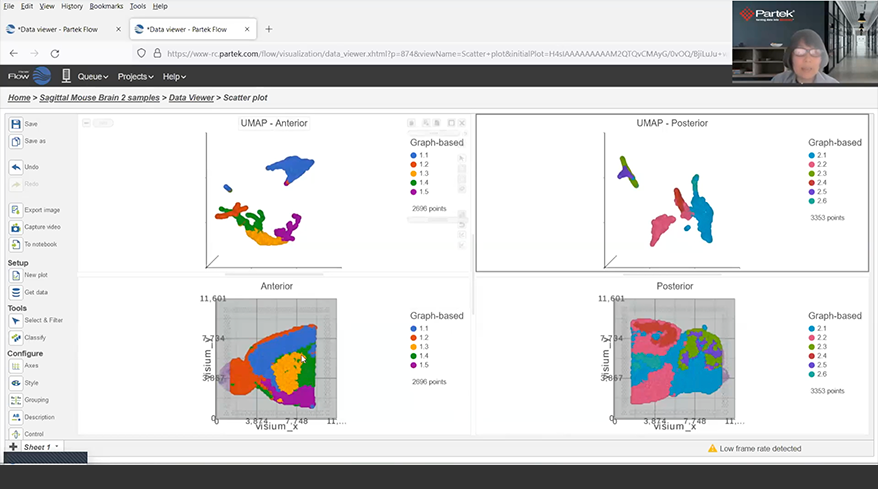
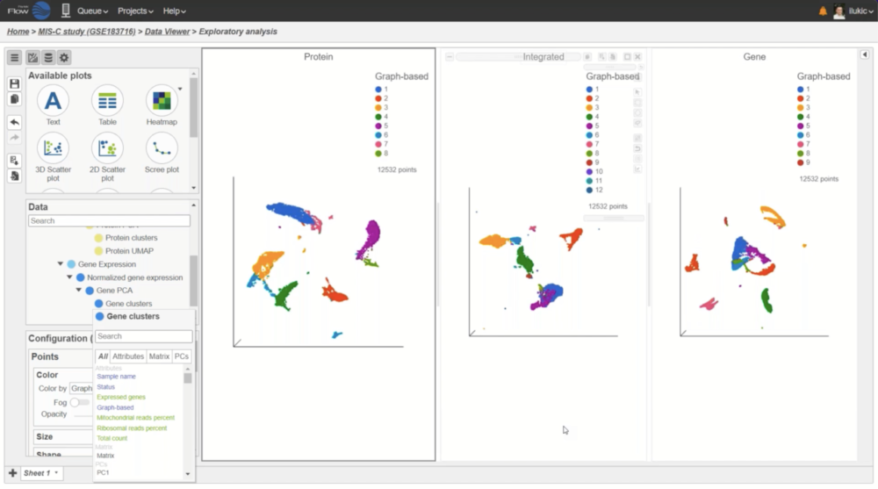
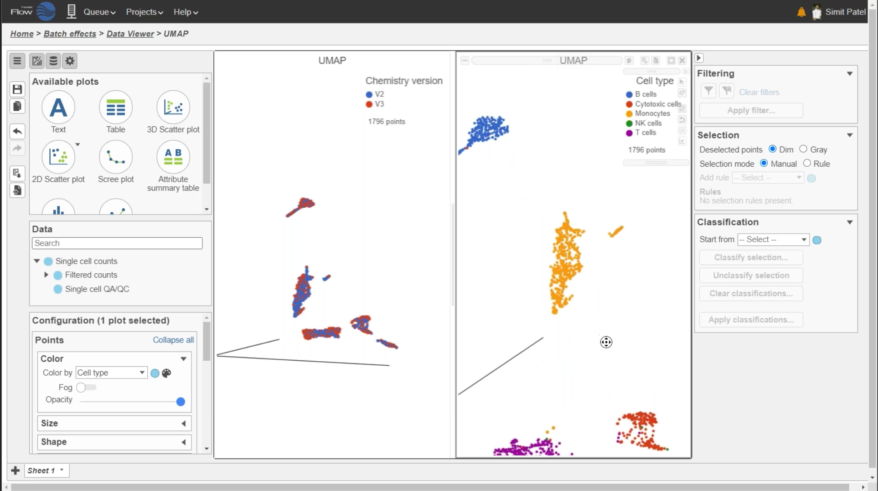
- New Harmony and Seurat 3 integration methods for batch removal
- New options for Descriptive Statistics
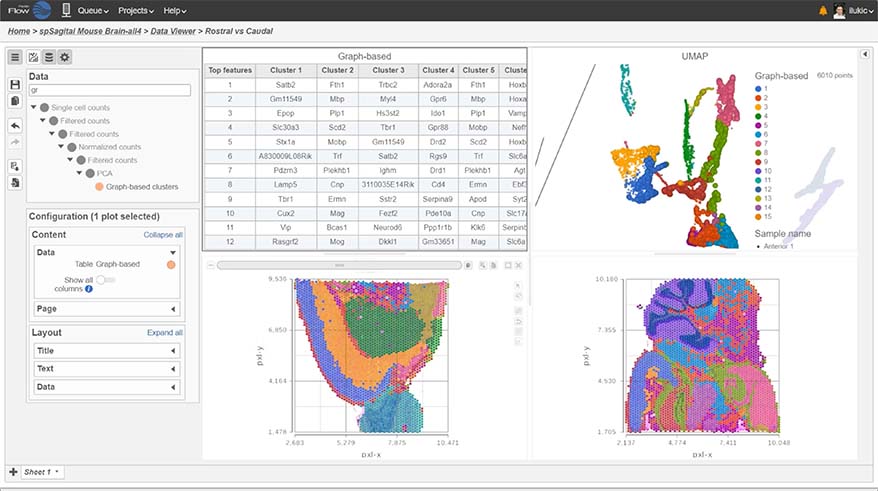
- New similarity matrix
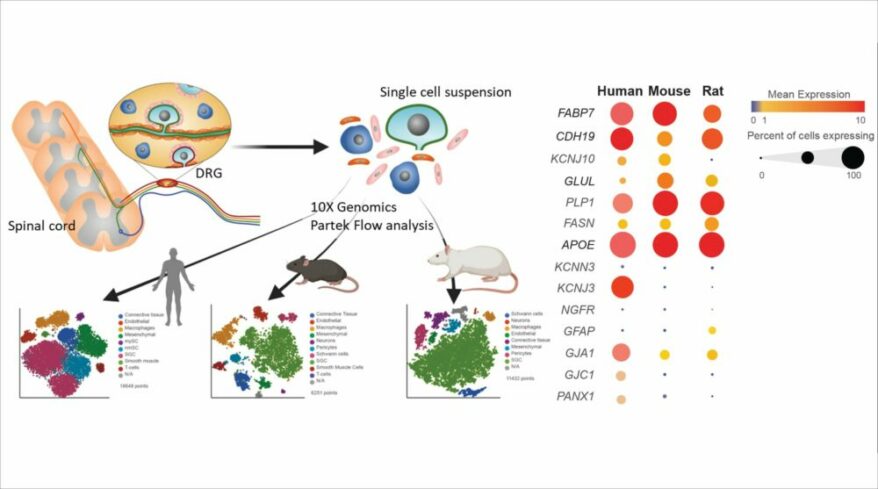
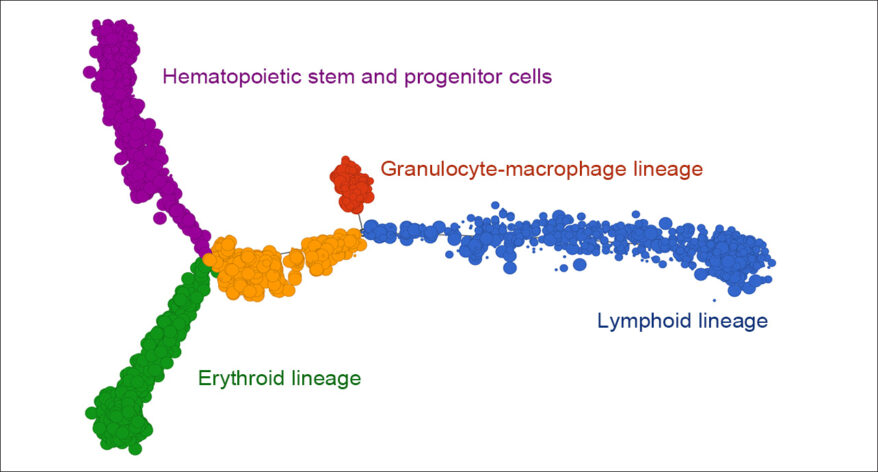
- New Biological interpretation of miRNA-Seq data
- And more!
Register to Watch
Our Single Cell Webinar Series
Share this post: