Whole genome, exome, or targeted panel DNA sequencing (DNA-Seq) studies can identify genetic variants that contribute to disease and phenotypes. Partek offers powerful and easy-to-use tools for processing and analyzing your DNA sequencing data. With the latest databases and bioinformatics tools available in a point-and-click interface, variant analysis is simple.
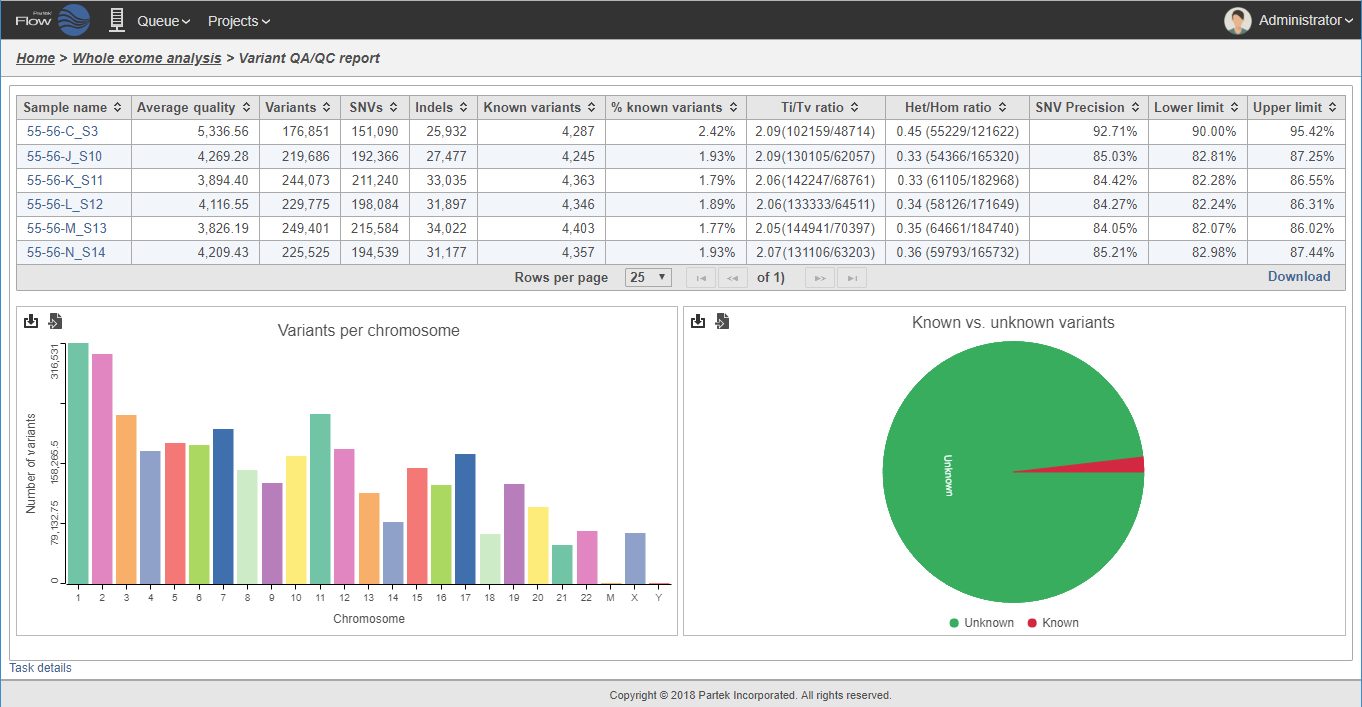
Inspect and explore your variants using informative reports.
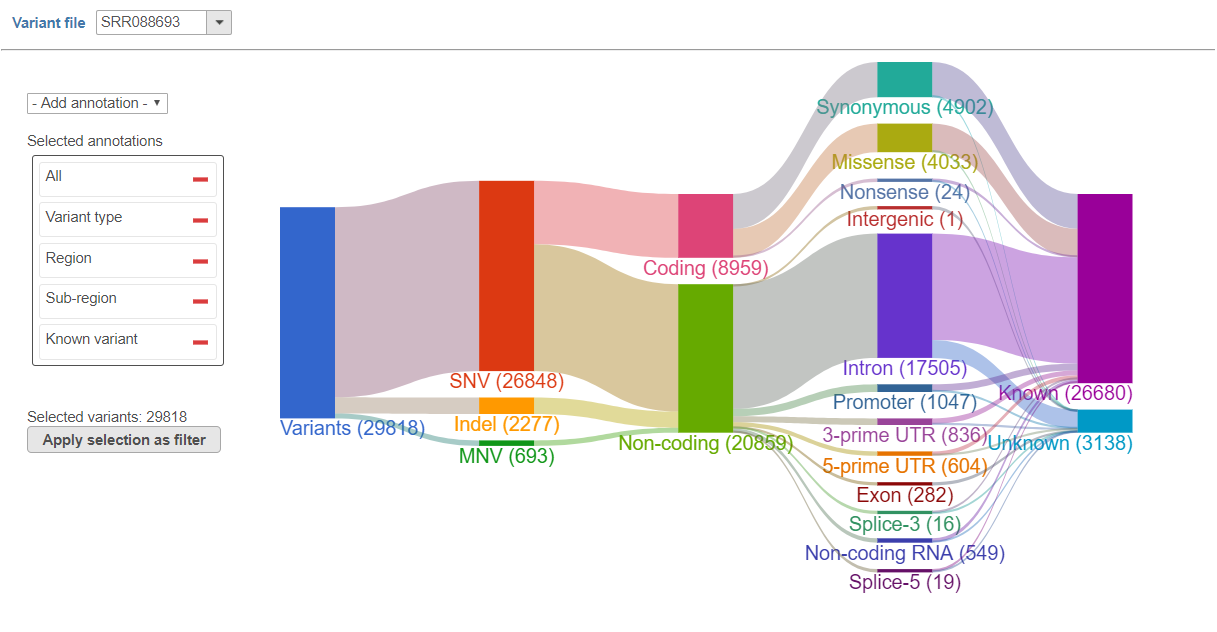
Detect somatic and germline variants using the latest algorithms.
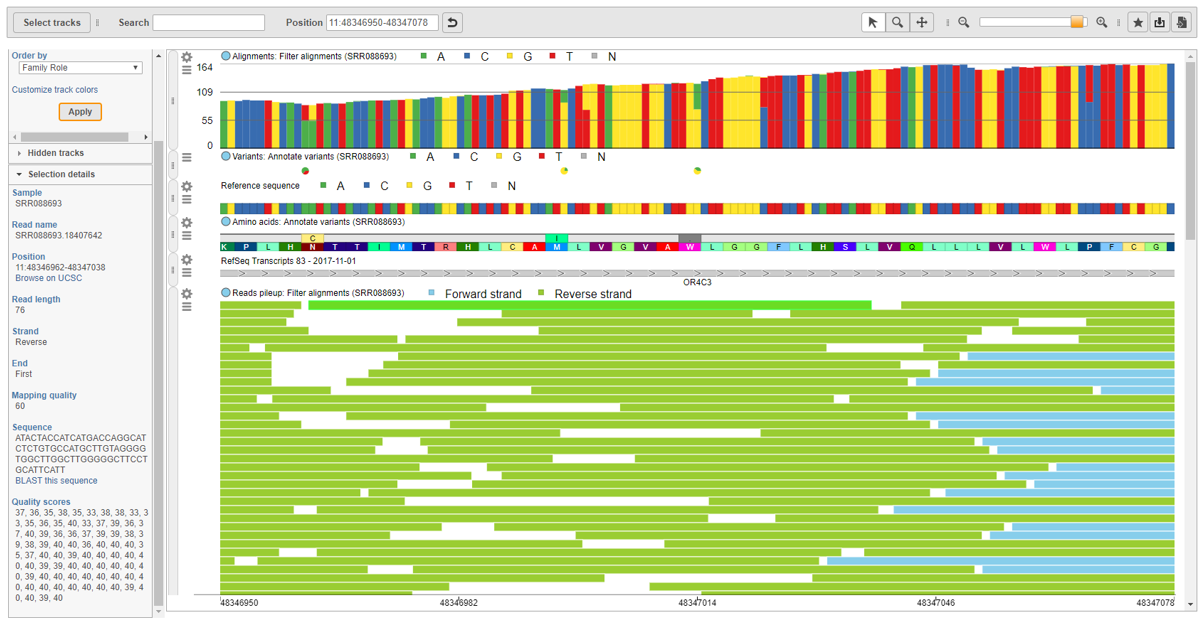
Visualize aligned reads and variant calls using the interactive genome browser.

QA/QC

Alignment


Detect Variant


Annotate Variants


Filter Variants

Analyze Your Variant Detection Data with Confidence
To detect variants with confidence, you must know that your sequencing data is of high quality. At each step in the process from raw data to variant calls, Partek Flow provides comprehensive QA/QC reports to help you check your data quality and tools to remove low-quality data.
For FASTQ files, the pre-alignment QA/QC report details each sample so that you know you are working with high-quality data. If you do need to remove low-quality reads or base calls, Partek Flow has easy-to-use quality trimming and read filtering tasks to help you clean up your data.
Aligning reads is easy using Partek Flow. We provide human, mouse, and rat references, but also make it easy to add a reference genome for any species. We offer fast and accurate aligners for DNA-Seq analysis, including BWA. Our post-alignment QA/QC report gives detailed information about each sample and lets you identify any technical issue before continuing with variant detection. To remove any PCR artifacts from your sequencing data, you can quickly filter out duplicate aligned reads.
Our coverage report helps you evaluate the sequencing depth of each sample and confirm that all samples or regions have sufficient depth for detecting variants.

All the Best Tools for Variant Detection in One Place
Partek Flow offers variant callers suitable for any variant detection study. Choose from GATK4 HaplotypeCaller, FreeBayes, LoFreq, and SAMtools for germline variant detection; GATK4 Mutect2 and Strelka for somatic variants; LoFreq for low frequency variants in cfDNA or ctDNA samples; and CNVkit for copy number changes.
A challenge in variant analysis is defining a final set of high-confidence variant calls. Partek Flow makes it easy to compare results from different variant callers. By combining calls from two different algorithms, you can identify variants that are reliably detected by different methods.
For any specific variant, the integrated chromosome viewer lets you visualize the variant in the context of the genome, view transcripts from multiple annotations, and examine details about individual reads to verify the variant call.
After your analysis is complete, Partek Flow makes it easy to download your VCF files from the Partek Flow server to your local machine.

Identify Causal Variants
Filtering from all detected variants to potentially causal variants is a critical step in variant analysis. Partek Flow makes this process easy, providing rich annotations about each variant and making the filtering process simple.
Using information from public databases, including 1000 Genomes and dbSNP, you can add information about the population frequency and disease associations of known variants. With the powerful predictive tools VEP and SnpEff, you can predict the functional impact of even novel variants on protein function.
Once your variants are annotated, Partek Flow makes it easy to filter variants using the interactive Sankey plot. Just point and click.

Selected Variant Detection Publications Citing Partek Software
Multiomics Characterization of Low-Grade Serous Ovarian Carcinoma Identifies Potential Biomarkers of MEK Inhibitor Sensitivity and Therapeutic Vulnerability
Shrestha, Raunak et al., Cancer Research (2022)
Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target
Fernandez, Llaurado Marta et al., Cancer Cell International (2019)

Ask a question or request a software demonstration/trial
