Chromatin immunoprecipitation sequencing (ChIP-Seq) is used to determine how proteins interact with DNA to regulate gene expression. Partek provides easy-to-use tools that guide you through the entire analysis process from start to finish.
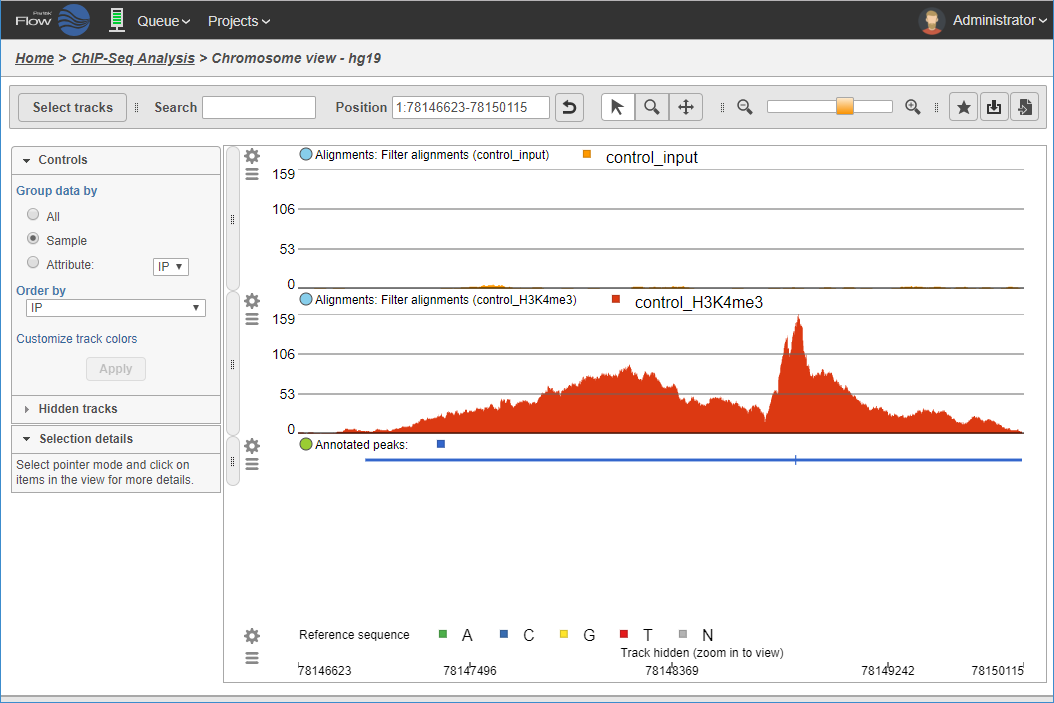
Visualizing aligned reads in ChIP sample and control sample, with detected peak region track.
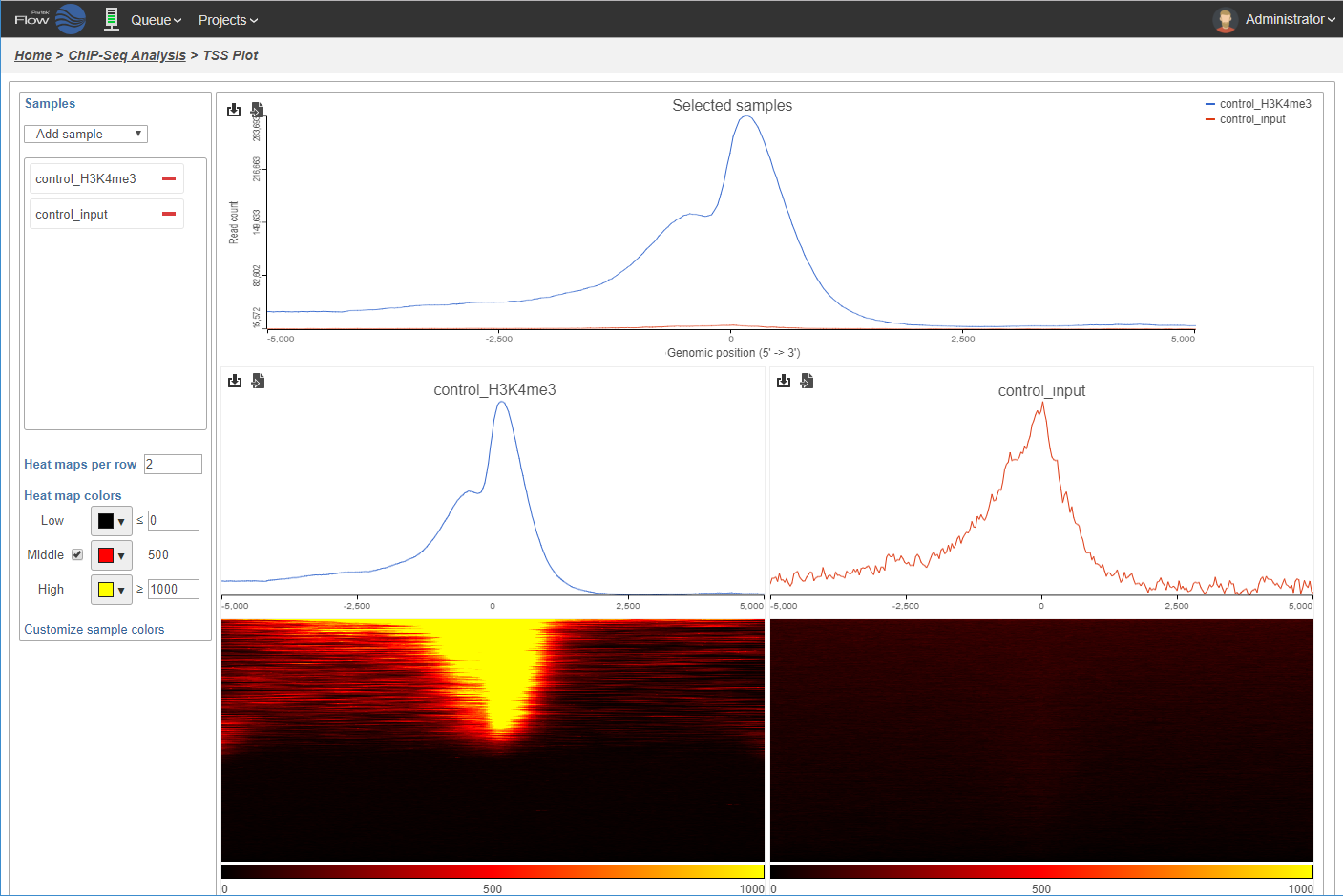
TSS plot displays the read coverage around the transcription start sites within the detected peaks in ChIP sample.
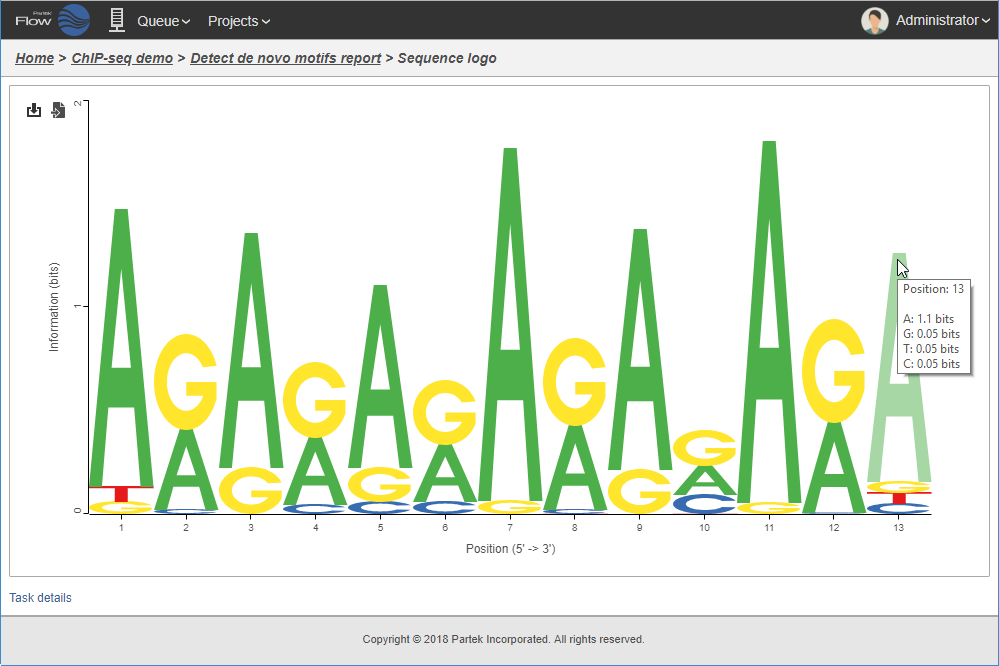
Logo view of motif detected in the binding sites.

Alignment

Peak Detection


Peak Annotation


Motif Detection

Visualization

Fast and Easy ChIP-Seq Analysis
ChIP-Seq is a powerful method to detect genome-wide DNA binding sites for transcription factors and other proteins. Many tools are available on the market to detect peaks and discover motifs from peak sequences, but most are command-line based. Partek provides a point-and-click interface making it fast and easy to build a start-to-finish ChIP-seq pipeline. Pipelines can also be easily built for other genomic locus analysis experiments such as ATAC-Seq, MNase-Seq, DNase-Seq, and more.

Integrate ChIP-Seq Data with Other Assays
Transcription factors (TF) control regulation of expression. Partek software allows you to easily integrate ChIP-Seq and RNA-Seq studies into a single study. Once peaks and TF binding sites are identified in ChIP-Seq data, peaks can be annotated to the genome to find the nearest transcription start site. A list of target genes generated from ChIP-Seq data can be compared with a list of differentially expressed genes generated from RNA-Seq data using a Venn diagram. The Partek chromosome view allows you to visualize the peaks and differentially expressed genes together.
Selected ChIP-Seq and ATAC-Seq Publications Citing Partek Software
GATA6 defines endoderm fate by controlling chromatin accessibility during differentiation of human-induced pluripotent stem cells
Heslop, James A. et al. Cell Reports (2021)
Genomic landscape of transcriptionally active histone arginine methylation marks, H3R2me2s and H4R3me2a, relative to nucleosome depleted regions
Beacon, Tasnim H. et al. Gene (2020)
Molecular mechanisms of Guadecitabine induced FGFR4 down regulation in alveolar rhabdomyosarcomas
Darvishi, Emad et al. Neoplasia (2020)

Ask a question or request a software demonstration/trial

