Copy number variation (CNV) is a type of structural variation and it encompasses duplication or deletion events that affect a considerable section of the genome. Those events may be mirrored by changes in gene expression levels and are consequently involved in pathogenesis. Partek tools enable you to start with raw data, detect amplified or deleted regions, and interpret or integrate the data with other assays such as gene expression.
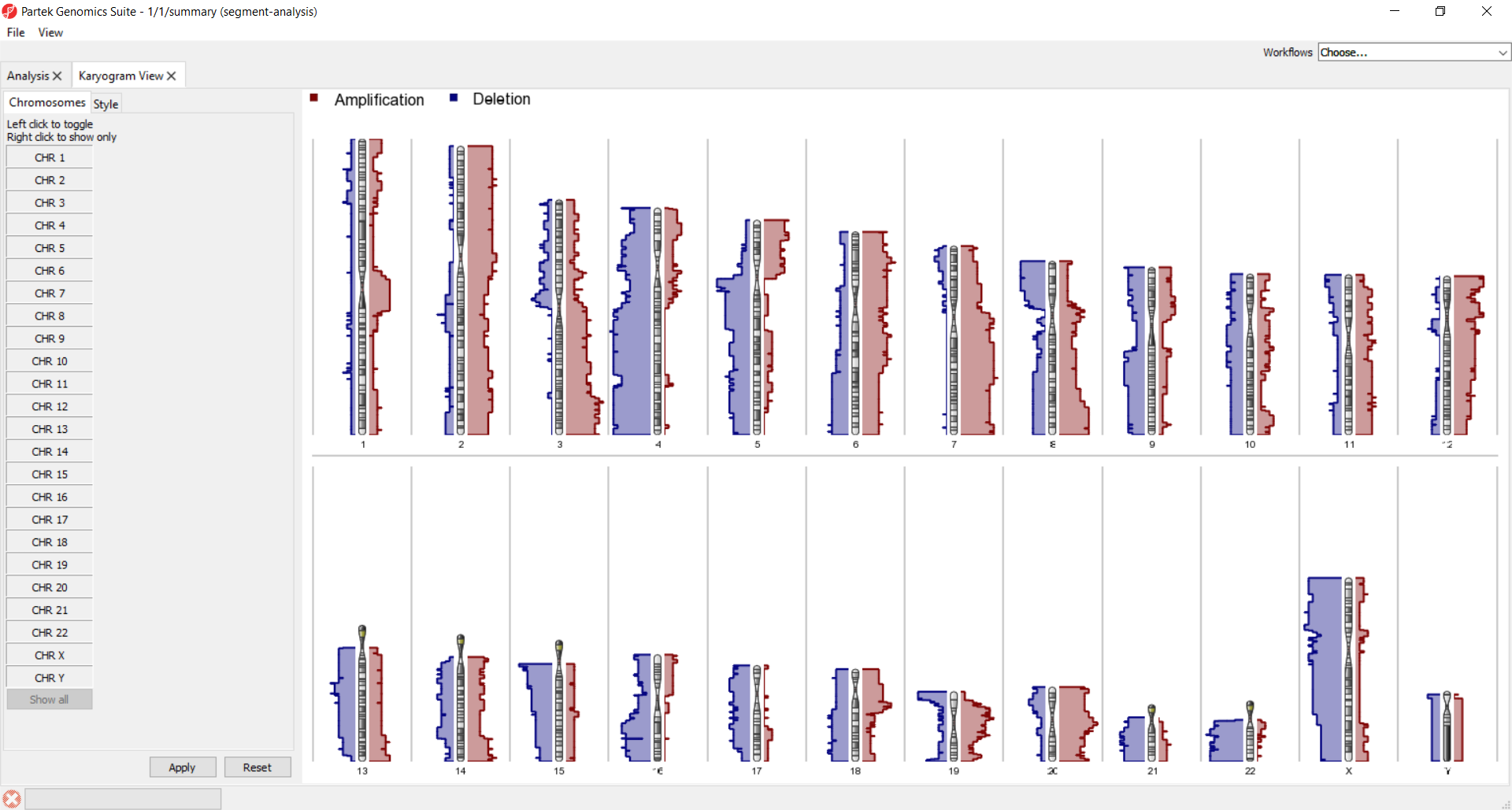
Summarize copy number changes by karyogram view.
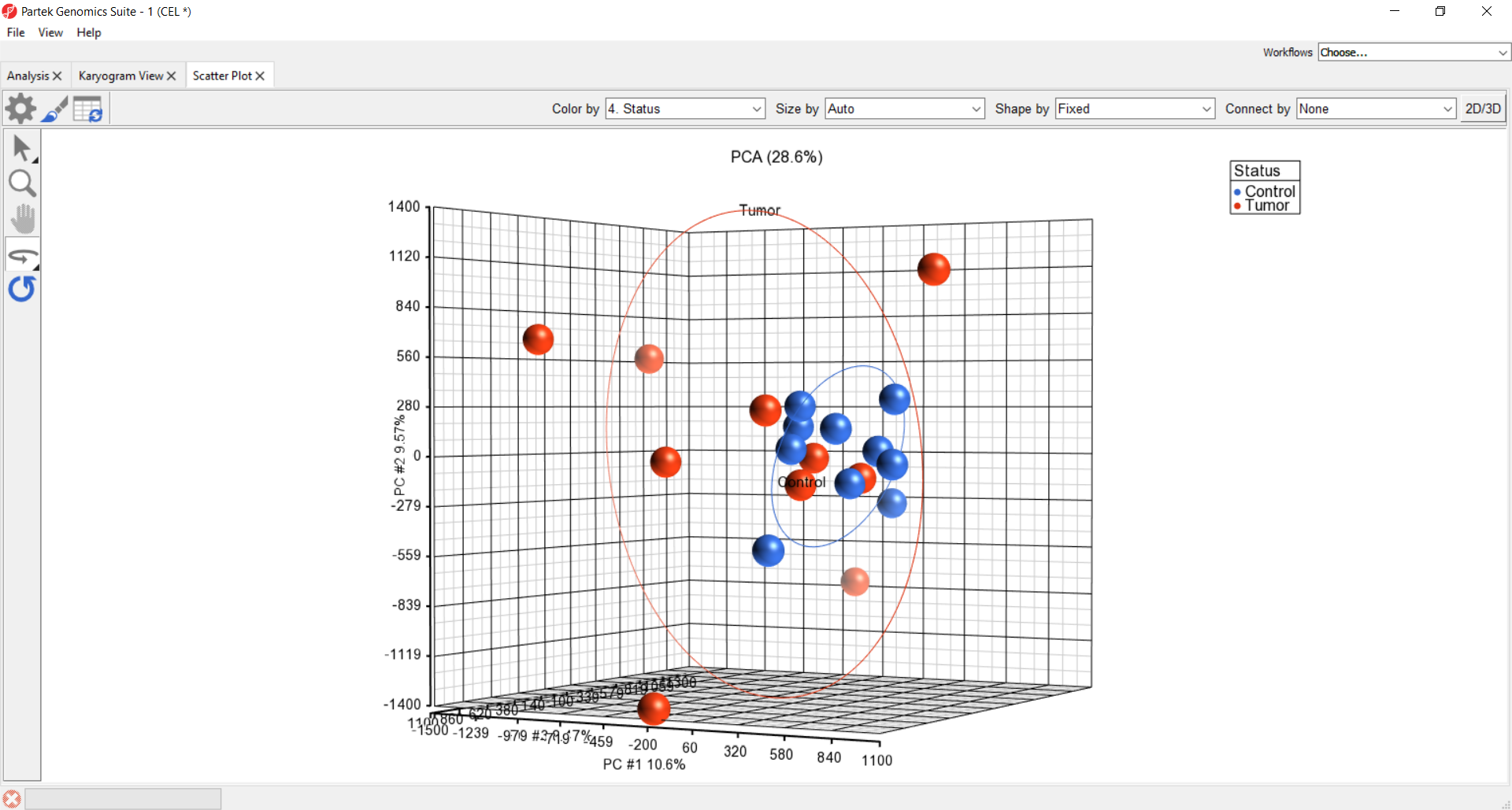
Explore copy number patterns by principal components plot.
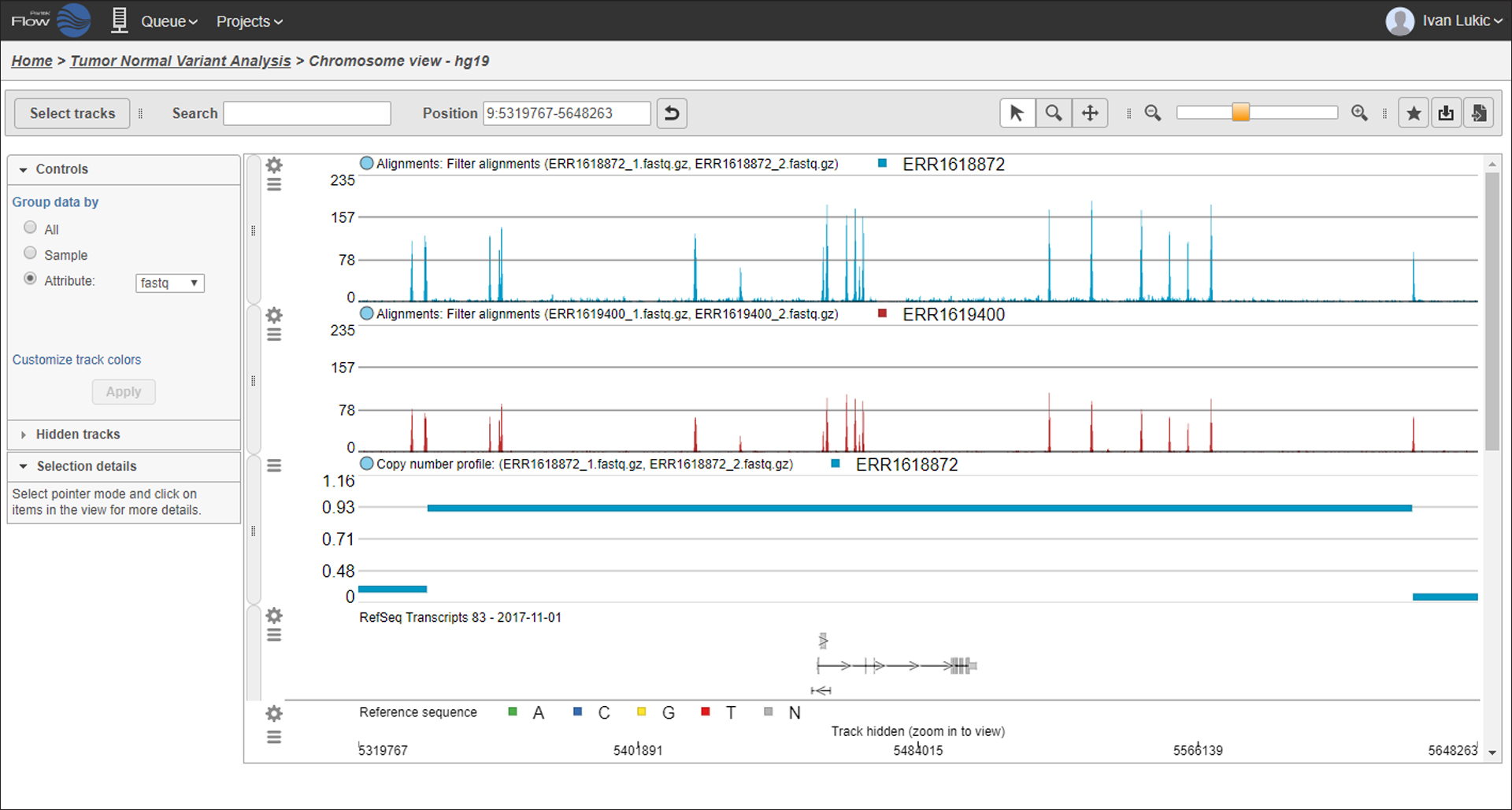
Visualize amplified or deleted regions.

Detect Amplifications or Deletions

View Detected Regions


Find Overlapping Genes

Biological Interpretation

What are Copy Number Variations?
Copy number variations are used to describe the differences in the number of copies of genes from one individual to another. This structural variation can result in some regions of the genome duplicating, triplicating, or even quadruplicating in some people. If these genomic regions include gene-rich regions, then this might mean some individuals will have four or more copies of a particular gene instead of the usual two copies. How copy number variations contribute to disease is an area of intense research. Diseases such as cancer are observed to be associated with increases in copy number of certain genes. Next generation sequencing is beginning to uncover the consequences of some of these copy number variations and how they relate to pathogenesis.
Pathways Affected by Copy Number Alterations
One of the major aims of copy number analysis is to find the genes that are located within amplified or deleted regions. Obtaining a list of the genes is the first step and the next is to interpret the list by identifying overrepresented gene groups such as pathways or gene ontology categories.
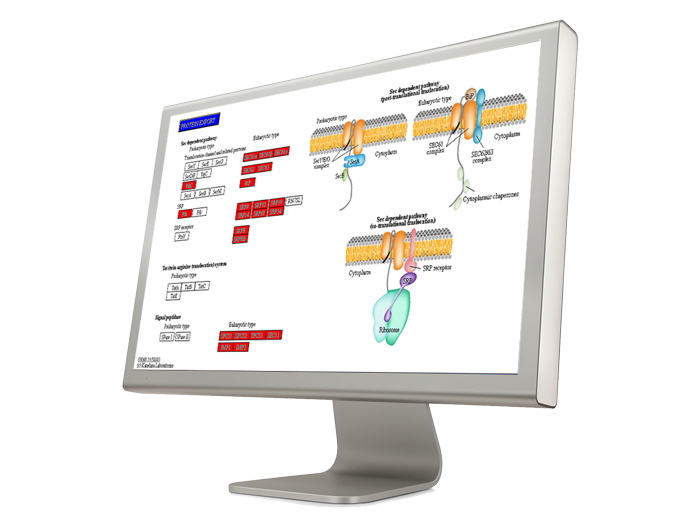
Integrating Copy Number and Gene Expression Analysis
Since copy number alterations can alter gene expression, an intriguing possibility is to combine the results of the two assays and look for evidence that gene deletions are mirrored by diminished gene expression levels. Partek tools enable you to integrate the two sets, use advanced statistics to substantiate your findings, and depict the results.
Impact of CNV Analysis in Medical Research
CNV analysis can help identify genes underlying common diseases. It can also help to fish out chromosomal rearrangements underlying developmental disorders. Identification of CNV’s that lead to either susceptibility or disease resistance can help in targeting these genomic areas in a more precise manner. The tools provided by Partek Flow for CNV analysis facilitates these kinds of discoveries.
Ask a question or request a software demonstration/trial
