Multisample Spatial TranscriptomicsAnalysis Without CodingPoint, Click, and Done Using Partek Flow
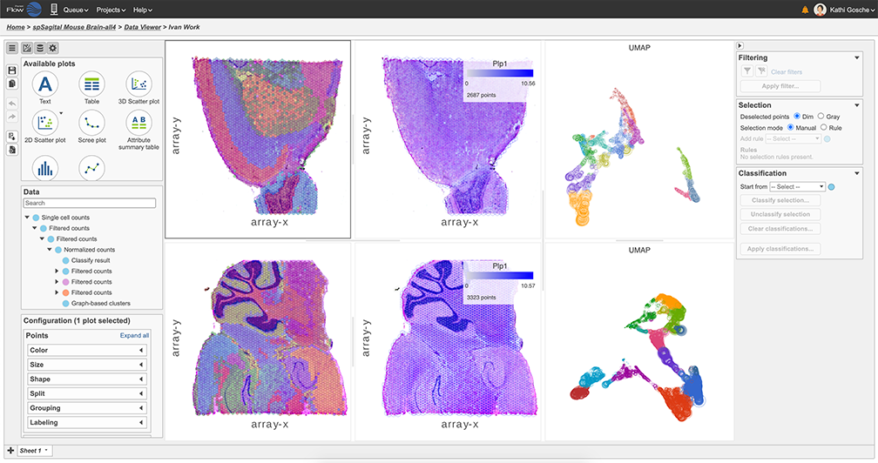
Spatial transcriptomics has revolutionized the field of gene expression analysis by bridging the gap between genomic information and tissue architecture. Partek® Flow® bioinformatics software, which is available to Genomics WA researchers, simplifies the exploration and analysis of transcriptomic data with an easy-to-use point-and-click interface.
Join us as we explore two human prostate samples, comparing healthy cancerous cells to identify differentially expressed genes and potential tumor biomarkers.
Agenda:
- QA/QC, filtering, and normalization
- Dimensionality reduction, unsupervised clustering, and cancer biomarker discovery
- Downstream biological interpretation to make comparisons and perform differential analysis
- Interactive visualization of spatial data
