How to Analyze Single Cell RNA-Seq Data with the inDrop System and Partek Flow
co-sponsored by

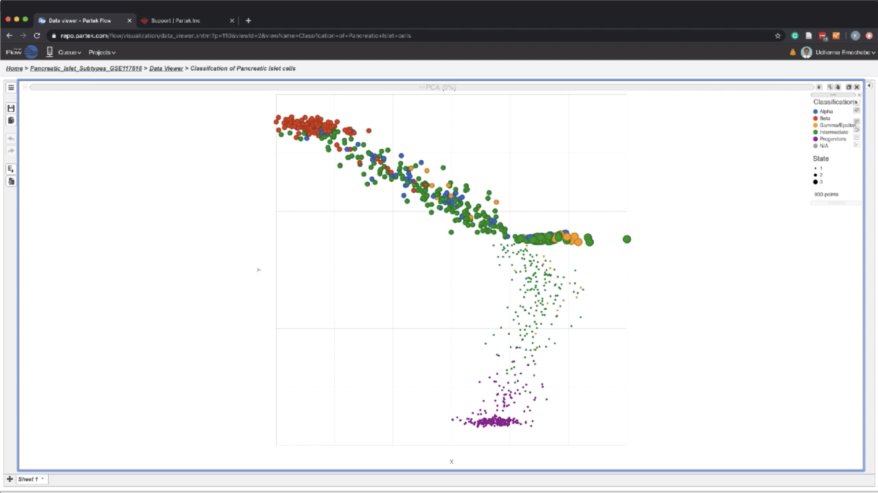
Single cell mRNA sequencing allows for the identification of different cell subtypes in a progenitor population. During pancreatic development, Neurog3 positive cells are destined to become endocrine cells generating alpha cells, beta cells, and delta cells. However, single cell data analyses can be challenging for those without programming or command line experience. Partek® Flow® bioinformatics software resolves this challenge with a simple and intuitive graphical interface that doesn’t sacrifice flexibility or statistical power.
In this webinar scientists from 1CellBio and Partek discuss how you can use the inDrop™ platform and Partek Flow to simplify single cell RNA-Seq analysis. There is a live demonstration of Partek Flow using an inDrop single cell mRNA sequencing dataset.
You will learn how to:
- identify cellular subtypes of Neurog3 positive cells
- use information-rich and interactive visualizations to identify graph-based clustering and cluster classification
- perform trajectory analysis of Neurog3 positive cells
