Recently a post about how the microscopic water bear (tardigrade) can survive in extreme drought, temperature, pressure, and radiation as well as in the vacuum of space popped up on my Twitter feed. Being the fan I am of anything space-related I was intrigued. I downloaded the publicly available data to explore what is it about the water bear transcriptomic profile that allows them to survive the extreme environmental difference between Earth and space. This presented the perfect opportunity for me to see how well Partek Flow software would let me analyze non-model organism data, and at the same time, not have to deal with any command-line tools.
Water bears survive by shriveling up and entering a dormant state, called the ‘tun’ state, so I compared samples from tun and active adults. I started by aligning the RNA-Seq data to the water bear reference genome (in fact, I could use the reference genome of any model or non-model organism in Partek Flow). To improve the quality of the analysis, I used the visualizations in the post-alignment QAQC and quantification reports to help identify a low-quality sample and remove it from my downstream analysis (Figure 1 and Figure 2).
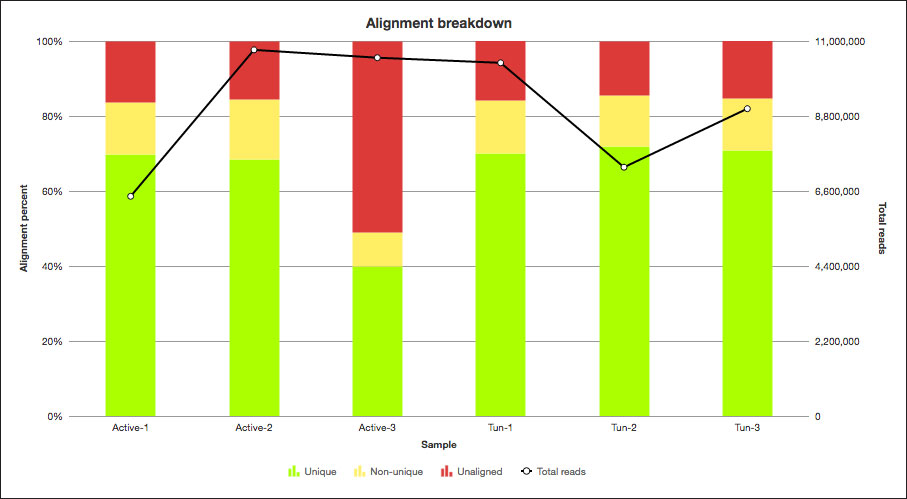
Figure 1: Post QAQC alignment breakdown of a non-model organism.
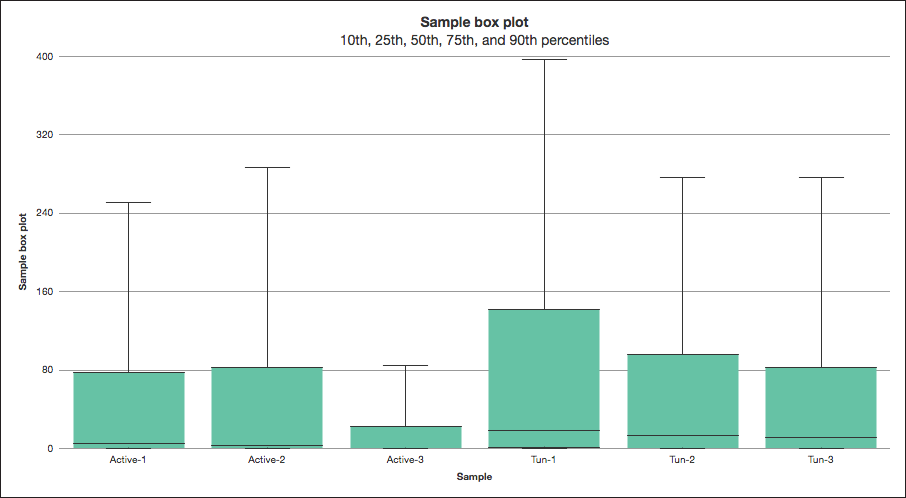
Figure 2: Quantification log expression signal of a non-model organism
I found 1385 genes that were differentially expressed between tun and active adults (Figure 3). Interestingly, I found most genes (1194 genes) were up-regulated in the tun group, compared to the active adults, indicating the transformation into the tun state requires the activation of many genes, rather than inactivation. This was rather surprising. When in the tun state, the water bears seemingly regress into dormancy. Intuitively, one would expect more inactivation. Then again, the transformation process requires major physiological, cellular, and metabolic changes, which would require the activation of pathways that would normally not be needed when they are active adults. Biological interpretation of the gene list revealed functions involved in oxidative damage response, redox reactions, and membrane integrity, all of which have been implicated in extreme stress response.
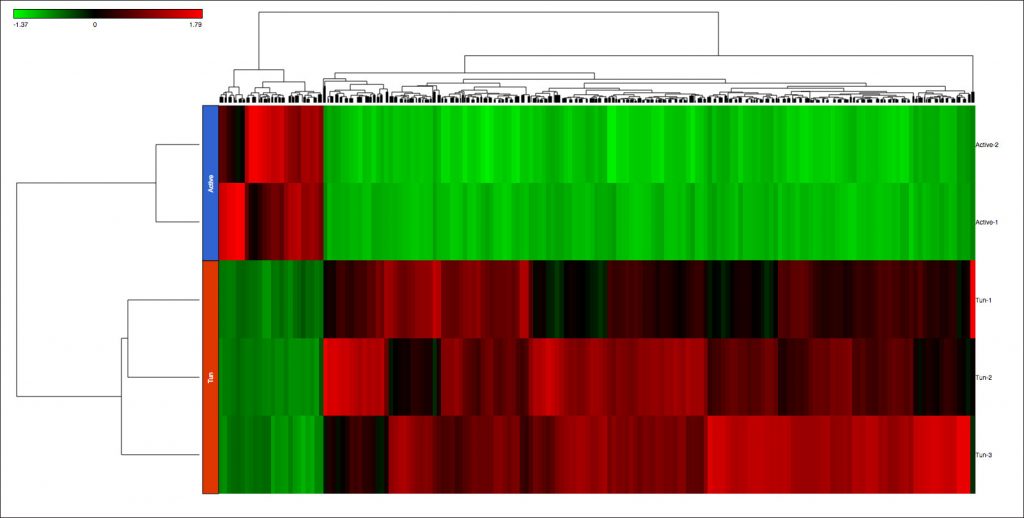
Figure 3: Dendrogram of 1385 genes that were differentially expressed between tun and active adults
The whole analysis, from data download to biological interpretation of the gene list, was completed in a matter of hours with a few easy clicks thanks to the intuitive graphical user interface in Partek Flow. Want to see how I did it? Watch the full demonstration in my webinar How to Analyze RNA-Seq Data From a Recently Sequenced Non-model Species of Water Bear Using Partek Flow.
References:
- Arakawa, K. et al. Genome Sequencing of a Single Tardigrade Hypsibius Dujardini Individual. Sci. Data 3:160063 doi: 10.1038/sdata.2016.63 (2016)
- Koutsovoulos, Georgios et al. “No Evidence for Extensive Horizontal Gene Transfer in the Genome of the Tardigrade Hypsibius Dujardini.” Proceedings of the National Academy of Sciences of the United States of America 113.18 (2016): 5053–5058. PMC. Web. 16 Nov. 2016.

