How to Analyze Lexogen QuantSeq Data with Partek Flow
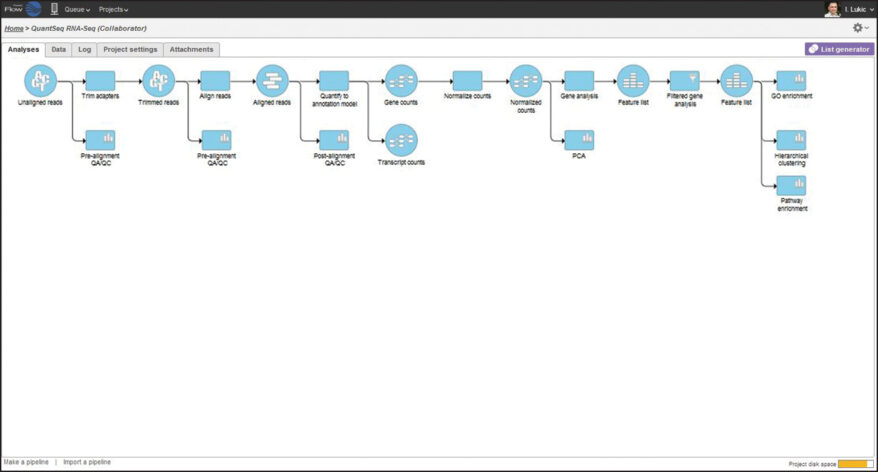
The Lexogen QuantSeq expression profiling library prep kits enable fast, easy, and cost-effective sequencing by gene counting and are an exceptional alternative to standard RNA-Seq protocols. Using the QuantSeq 3′ mRNA-Seq pipeline in Partek Flow, QuantSeq data processing is fast and easy. By combining powerful statistics and interactive visualizations in an intuitive graphical user interface, Partek Flow helps you get the most biology out of your data.
In this webinar, Lexogen and Partek scientists will show you how quick and easy it is to go from raw QuantSeq data to biological insights using the QuantSeq pipeline in Partek Flow.
Register to Watch
Share this post:
