Gene expression is the most fundamental level at which the genotype gives rise to a phenotype. Whether you have RNA-Seq, qPCR or microarray data, Partek provides easy-to-use tools that guide you through the analysis process from start to finish within a point-and-click interface. With over 9,000 citations, see why our gene expression analysis tools are the most trusted in the genomics community.
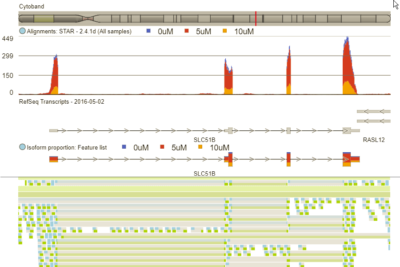
The viewer can display aligned reads, genes, transcripts, variants, isoform proportions, and reference sequence.
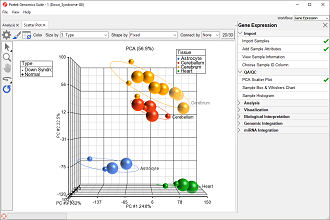
Principal components (PC) analysis (PCA) is an exploratory technique that is used to describe the structure of high dimensional data by reducing its dimensionality.
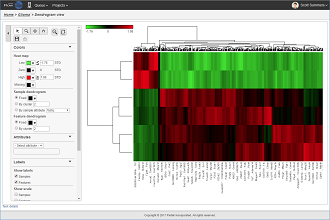
Heat maps are well-suited for visualizing large amounts of multi-dimensional data and can be used to identify clusters of rows with similar values.

Alignment

QA/QC


Quantification

Differential Analysis

Biological Interpretation

Powerful Multivariable Stats for Gene Expression Data
Multiple experimental variables can impact gene expression. However, a vast majority of analysis tools available cannot be configured to account for multiple factors. Partek tools can handle even the most complicated experimental designs. With our flexible filtering options, it is easy to remove technical noise and focus on the genes that matter in your experiment.
Starting with raw data, we give you the power to obtain meaningful results with confidence. You can even use widely-accepted methods to normalize your data, including total count, RPKM, and TMM for RNA-Seq data.

Get Meaningful Results
There is more to gene expression analysis than finding differentially expressed genes. With Partek tools, you can go beyond a list of genes to interpret the changes you find in the context of biological pathways and gene sets using KEGG pathway and Gene Ontology term enrichment analysis.
Partek tools can dig deeper into RNA-Seq data to identify alternatively spliced transcripts between experimental conditions. Even better, the integrated genome browser makes visualizing the isoform expression results fast and easy.
Partek software can help you answer complex experimental questions. Whether you are looking for patterns of gene expression change in time series data, building a classification model from published data, or discovering biomarkers by finding correlation between genes and clinical outcomes, powerful Partek tools enable and simplify your analysis.
When planning your next study, Partek tools are there to help. With our power analysis feature, you can design a more effective experiment by identifying how many samples you would need to measure a specific effect.
Integrate Gene Expression with Other Genomic Datasets
Regulatory elements and genetic variation can drive differences in gene expression. Analyzing multiple genomic datasets in parallel is a powerful way to identify molecular mechanisms underlying phenotypes.
A major bottleneck for integrated genomics studies is combining the data and results from different assays. Partek makes this data integration simple with tools to combine gene expression data from RNA-Seq or microarray studies with a wide variety of assays, including ChIP-Seq, Copy Number Variation (CNV), methylation, and microRNA expression.
Selected Gene Expression Publications Citing Partek Software
Human engineered meniscus transcriptome after short-term combined hypoxia and dynamic compression
Szojka, Alexander RA et al., Journal of Tissue Engineering (2021)
Effects of High-Fructose Diet vs Teklad Diet in the MNU-Induced Rat Mammary Cancer Model: Altered Tumorigenesis, Metabolomics and Tumor RNA Expression
Kumar, Amit et al., Journal of Obesity and Chronic Diseases (2021)
Multiomics analyses of cytokines, genes, miRNA, and regulatory networks in human mesenchymal stem cell expanded in stirred microcarrier-spinner cultures
Tin-Lun Lam, Alan et al., Stem Cell Research (2021)
Gene expression in the epileptic (EL) mouse hippocampus
Lee, Tih-Shih et al., Neurobiology of Disease (2021)

Ask a question or request a software demonstration/trial

